
Social Network Analysis for Computer Scientists
This is the old course page! See the new page for recent information!
This course deals with computer science (CS) aspects of social network analysis (SNA), and is open to all students in the master computer science programme at Leiden University.
In the Fall 2020 semester, Social Network Analysis for Computer Scientists is a hybrid on-site/online course.
Update 18-12-2020: Final grades have been submitted to the student administration.
Subscribe to the Online and Lab Session Live Room (one-time action):
https://video.leidenuniv.nl/channel/SNACS+2020/177222361/subscribe
Subscribe to the On-site Live Room (one-time action):
https://video.leidenuniv.nl/channel/SNACS+2020+-+On-site+room/185235803/subscribe
(most links require ULCN login)
Course information
Lectures: Fridays from 8:30 to 10:15
in Gorlaeus room 03 (online; presenters on-site: Snellius 174 or online)
Lab sessions: Fridays from 10:30 to 12:15
in Gorlaeus room 03 (online)
Prerequisites: a CS bachelor with courses on Algorithms, Data Structures and Data Mining
Literature: provided papers and book chapters (free and digitally available)
Examination: based on presentation, paper, programming, peer review and participation (no exam)
Brightspace link: 2021-WN Social Network Analysis for Computer Scientists
Study points: 6 ECTS
Lecturer: dr. Frank Takes - f.w.takes@liacs.leidenuniv.nl, room 157b
Assistant lecturer: Anna Latour MSc - a.l.d.latour@liacs.leidenuniv.nl, room 123
Assistants:
Antonio Barata MSc - a.p.pereira.barata@liacs.leidenuniv.nl, room 126
Hanjo Boekhout MSc - h.d.boekhout@liacs.leidenuniv.nl, room 126
Jingsen Chen - j.chen.22@umail.leidenuniv.nl
![[Network visualization image]](yeast_eccs.jpg)
Network with 1458 nodes and 1948 edges.
Course schedule
| Date | First slot (8:30-10:15) | Second slot (10:30-12:15) | |
|---|---|---|---|
| 1. | Sep 4, 2020 |
Lecture 0: Course information (video,pdf) Lecture 1: Introduction and small world phenomenon (video,pdf) |
Introduction to Gephi (video) Work on Assignment 1 |
| 2. | Sep 11, 2020 | Lecture 2: Advanced network concepts and centrality (video,pdf) | Introduction to NetworkX Work on Assignment 1 |
| 3. | Sep 18, 2020 | Lecture 3: Network projection and community structure (video,pdf) Presentation 0: Example course project presentation (video,pdf) |
Project team formation Work on Assignment 1 |
| 4. | Sep 25, 2020 | Lecture 4: Network structure of the web (video,pdf) | Work on Assignment 1 |
| Sep 28, 2020 | Deadline for Assignment 1 (hand in via Brightspace) | ||
| Sep 30, 2020 | Deadline for having chosen a course project topic in Brightspace (and reserved a presentation slot) | ||
| 5. | Oct 2, 2020 | Lecture 5: Network dynamics and evolution (video,pdf) | Introduction to data science lab & Course project planning Work on Assignment 2 |
| 6. | Oct 9, 2020 | On-site presentation 1: Visualization algorithms 1
On-site presentation 2: Link prediction 2 Online presentation 1: Link prediction 1 Online presentation 2: Community detection 1 |
Work on Assignment 2 |
| 7. | Oct 16, 2020 | On-site presentation 3: Shortest paths 1
On-site presentation 4: Influence spread and virality 1 Online presentation 3: Diameter computation Online presentation 4: Anomaly detection 1 |
Work on Assignment 2 |
| 8. | Oct 23, 2020 | On-site presentation 5: Closeness centrality 2 (top-k)
On-site presentation 6: Local cluster detection Online presentation 5: Community detection 3 Online presentation 6: Triangle counting |
Work on Assignment 2 |
| Oct 26, 2020 | Deadline for Assignment 2 (hand in via Brightspace) | ||
| 9. | Oct 30, 2020 | On-site presentation 7: Link prediction in signed networks
On-site presentation 8: Graph evolution rules Online presentation 7: Visualization algorithms 2 Online presentation 8: Sampling from networks 1 |
Work on course project paper |
| 10. | Nov 6, 2020 | On-site presentation 9: Betweenness centrality 2
On-site presentation 10: Community detection 4 Online presentation 9: Network embeddings 2 Online presentation 10: Influence spread and virality 2 |
Work on course project |
| Nov 13, 2020, 10:30 CET |
Deadline for having a first version of the course project paper ready in PDF for peer review by fellow students | ||
| 11. | Nov 13, 2020 | On-site presentation 11: Sampling from networks 2
On-site presentation 12: Graph compression Online presentation 11: Betweenness centrality 1 Online presentation 12: Motifs in multilayer networks |
Peer review session |
| 12. | Nov 20, 2020 | On-site presentation 13: Network embeddings 1
On-site presentation 14: Motifs in temporal networks Online presentation 13: Community detection 2 Online presentation 14: Visualization algorithms 3 |
Work on course project |
| Nov 23, 2020 | Optional deadline for preliminary course project paper for feedback (hand in via Brightspace) | ||
| 13. | Nov 27, 2020 | On-site presentation 15: Neighborhood approximation
On-site presentation 16: Data errors in networks 1 Online presentation 15: Closeness centrality 1 Online presentation 16: Personalized PageRank |
Code review session |
| 14. | Dec 4, 2020 | On-site presentation 17: Dense subgraph detection
On-site presentation 18: Privacy and anonymity in networks Online presentation 17: Shortest paths 2 Online presentation 18: n/a |
Work on course project |
| Dec 9, 2020 | Deadline for final course project paper (hand in via Brightspace) | ||
| Dec 16, 2020 | Retake deadline for assignments | ||
| Dec 18, 2020 | Course end. Grades are sent to student administration. | ||
| Jan 29, 2021 | Retake deadline for course project | ||
The main goal of this lab session is to ensure that everything is set for making serious progress writing the course project paper in the coming weeks.
Course project paper
Earlier, you have made a project planning.
One upcoming deadline is the first version of your paper for the peer review session (November 11).
- Template. If you have not yet done so, download the paper template and make sure you can compile it and understand how to use it. Remember to fill in the meta information.
- Related work. If you have not yet done so, spend some time at Google Scholar investigating a) what other papers exist on this topic, b) which relevant papers cite your paper, and c) what important references are presented in your paper. You will likely want to include some of these in your paper as well. At least make sure that you are aware of the major works that cite your paper, and that you reference some of these works in the Introduction when you sketch the context of your work.
- Project contribution. For the course project, you have to do something original. Ideally, this goes beyond the one paper that you were assigned, comparing techniques from multiple papers, for example comparing different algorithms or methods, using different validation metrics, or testing on (a) larger (number of diverse) datasets. Try to get this written down in the appropriate section(s) of your paper.
- Collaboration. How will you work together on the paper? Git? Overleaf? How will you share code, data and results?
- What is a good paper?. What defines a good paper? Discuss with your project partner to align your expections. When are you "done"? When is the paper "sufficient"? When is it "good"? "Excellent"?
- Writing. Make serious progress with writing the first 3 to 4 sections of your course project paper in the coming two weeks, so that you have something substantial for the peer review session.
- Planning. Look at the planning you made early October. Have you reserved time to review each other's texts? Are adjustments necessary?
As always: please ask course staff for help if you are still unsure about certain aspects of your project.
The main goal of this lab session is to get started with both the course project and with Assignment 2, and to get to know the data science lab.
How-to set up remote access to LIACS REL
Data Science Lab
The data science lab website provides necessary information and documentation.
Become familiar with the lab and how to run code on it, and how to place data within the lab. This may be handy for Assignment 2 and/or the course project. Remember:
- /home/sXXXXXX, your homedirectory ~, is for your own code,
- /local is for local storage on the current machine you are are on,
- /data is for storing data across data science lab machines.
Course project
Below are some topics you can discuss and investigate together with your project team partner.
- Project schedule. Have a look at the deadlines for the course project in the schedule on this website, and create a sensible planning with your team partner.
- Paper. Read your course project paper, and spend some time at Google Scholar investigating a) what other papers exist on this topic, b) which relevant papers cite your paper, and c) what important references are presented in your paper.
- Data repositories. Check out the data repositories of real (social) networks such as SNAP and KONECT and think of datasets suitable for your course project.
- Project contribution. For the course project, you have to do something original. Ideally, this goes beyond the one paper that you were assigned, comparing techniques from multiple papers, for example comparing different algorithms or methods, using different validation metrics, or testing on (a) larger (number of diverse) datasets. Write down in at most 200 words what you plan to do for your course project, and feel free to discuss this with the lecturer or an assistant for feedback. Also see the generic instructions for the Course Project.
Assignment 2
Get started with the practical part of Assignment 2.
The main goal of this lab session is to become familiar with NetworkX (a Python package to analyze networks for research purposes).
All relevant information on NetworkX can be found in the NetworkX online documentation.
For this lab session, you need a working python environment. For this, there are two options:
Setup and installation
- Use your own self-installed python environment, and choose your own editor and way of running code (via the command line, an IDE or an interactive notebook.
- Alternatively, you can use the university's remote SSH functionality to work on a student machine in the computer rooms or a remote computing server. Below are some instructions for running python remotely in the command line on the ISSC infrastructure.
Access. First, connect to the gateway sshgw.leidenuniv.nl. Then, navigate to a student computer (see the REL page on SSH access, only accessible internally for a list of computers), or hop on to the remote server remotelx.liacs.leidenuniv.nl. You will work with python in a separate environment so that you can install packages using pip.
Installation.. To use Python in your own environment, first go (or make) the directory where you will work in for the course. For example ~/Documents/SNACS
Next, create a virtual environment with the following command:
virtualenv -p /usr/bin/python3.6 snacsenv
You can activate this environment with the following command (this is necessary each time you want to use your environment):
source snacsenv/bin/activate
You can install the packages you want with the pip command:
pip install numpy matplotlib networkx
Running code from the command line. Now you should be able to run your code from the command line by typing
python3 scriptname.py
Bonus: If you want to use an interactive jupyter notebook instead, this is possible with the following commands:
pip install jupyter
jupyter notebook --ip=127.0.0.1 —port=8888
Instructions for today: Lab session on NetworkX
- Take some time to do the NetworkX tutorial.
- Have a look at functionality to read and write graphs from/to disk, and in particular learn how to import an edge list. Understand the input format and ways of including things like Weight and other attributes of edges.
- A lot of the common network metrics you may want to compute are implemented as NetworkX function or NetworkX algorithm. Become familiar with these, for example by computing measures such as degree assortativity, clustering, density, diameter and average distance.
- While at it, why not take a look at how NetworkX relates to other data formats?
- Try to load the network from the first lab session (small-gephiready.tsv) into NetworkX, and investigate some characteristic properties of the network, such as the degree distribution and distance distribution. Use theread_edgelistfunction and remember to select the correct separator (tab), and pay attention to the header of the file.
- Get the Epinions network from the SNAP repository. You may need to fiddle with the precise heading, but it is already in edge list format. Compute some common characteristics such as the degree distribution, and visualize them using appropriate figures of distributions, for example using Matplotlib.
- What are the differences between NetworkX and Gephi in terms of visualization and analysis capabilities?
Done? Proceed with Exercise 2 of Assignment 1.
Looking for a challenge? Check out these two alternatives (that you can also use instead of NetworkX throughout the course, if you prefer (but for which there is less help available)):
- Graph-Tool, a python graph analysis toolkit that leaves the hard computation to parallel (OpenMP) C++ code.
- SNAP, which is entirely written in C++ and has many interesting features.
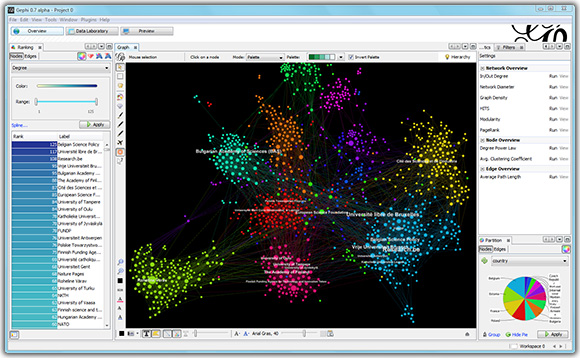
About social network analysis tools and packages. There exist different tools and package for social network analysis. In this course, we will introduce you to two of them, with complementary advantages:
- Gephi, an easy-to-use tool with a graphical interface useful for visualization
and quick analysis of relatively small network data (this week, see below).
Download Gephi 0.9.2 for Windows, Linux or Mac (mirrors provided because of occasional slow speed of gephi.org downloads). - NetworkX,
a more extensive Python package for network analysis that can handle larger network datasets and computations (next week).
Other even faster tools such as SNAP (in C++) can also be used, but require some more figuring out by the student.
Learning goals. The main goal of this lab session is to become familiar with Gephi (experimental beta-software to visualize networks for research purposes) and its input format. At the end of this session you should be able to:
- Know how to use Gephi for social network analysis, in particular
- Import and visualize raw network data with labeled nodes and labeled and/or weighted edges (directed or undirected),
- Understand how to map edge and node size and color to structural network properties such as the node degree and edge type,
- Know how to apply filters to the visualization, for example to focus only on the giant component,
- And export a vector graphic PDF of your network,
- Export computed node data for reuse in another program.
There is no deliverable for this lab session, but you are assumed to know the tool afterwards. Practice more afterwards at home if needed.
Instructions for today: Lab session on Gephi.
The following steps and tutorials will help you get to know Gephi.
- Install and run Gephi locally on your computer. Installation instructions, if needed, can be found on the Gephi website.
Note: If Gephi does not immediately work, you may have to adjust one line in etc/gephi.conf so that Gephi can find the right Java version (and remove the # in front of that line), reading something like jdkhome="/usr/lib/jvm/java-8-openjdk-amd64/" (or the equivalent Windows path). Also, Windows users sometimes have to launch the gephi64.exe instead of gephi.exe (or the other way around, depending on Java versions). - Walk through the Gephi tutorials. The Gephi website has various official tutorials that you can walk through; in particular Quick start, Visualization and Layouts. You may benefit from using a more modern tutorial (courtesy of Derek Geene at UCD).
Note: One reason to use a more modern tutorial is that the official Gephi Quick Start Guide is for a slightly older version of Gephi. On p. 9-10, contrary to what is indicated in the getting started guide, the ranking module is part of the appearance module (since Gephi 0.9), and does not appear as a separate tab. The “Ranking result” table no longer exists, and you can skip that part of the tutorial. In the tutorial on Visualization, on p. 9 the 3-D visualisation option was bugged and has been removed in Gephi 0.9. You can skip that part of the tutorial.
- Compute relevant real-world network dataset properties. Try out some of the Gephi datasets and see if you can understand how they differ in terms of density, degree distribution, giant components, clustering and average distance.
- Walk through a tutorial on data import and export. Learn how to import raw data from this short data import tutorial. Visualize the network represented by the edge list small-gephiready.tsv.
Compute the degree for each node and export it to a node list file (which you can for example reuse in other plotting tools).
- Visualization and exporting as vector graphic. Play around with some different visualization algorithms such as ForceAtlas 2 (for example, with scaling set to a higher value and with stronger gravity checked) and Fruchterman Reingold. Try to visualize the network such that labels remain readable.
Hints: From a random initialization, use the "ForceAtlas 2" visualization algorithm perhaps with "Stronger gravity" and usually with "Scaling" set to a much higher than default value. Increase the overall node size if needed. Once stable, you can improve the layout by enabling parameter "Prevent overlap". Run "Expansion" to make some more space between the nodes, then enable the display of node labels, properly size them, and finally run "Label Adjust" to prevent label overlap.
Export your visualization to PDF so that you have it as a vector graphic.
- Create, import and export your own social network data. Make a new, very simple, edge list input file of a fictive network with say 10 nodes and 20 edges, and import it into Gephi. Now add to the edge input file (in some order) link (un)direction, link weights and link labels.
Also import a node list file with node labels and other properties. Be sure to append it to your existing workspace and to use identifiers.
For example, have nodes represent students, and link represent friendships, and choose node and edge labels accordingly. Make sure that you are able understand the difference in file input and visualization output, and that you are comfortable with importing and exporting data into and from Gephi.
Done? Get started with the practical part of Assignment 1. You can download the smaller datafiles medium.tsv and large.tsv. If you want to analyze huge.tsv, you will have to get it from the shared folder stated in the assignment.
Course project
Teams work on a course project for 60% of the course grade. The project is about a certain topic related to social network analysis, and the project consists of:
- Giving a max. 30-minute presentation (+15 minutes for questions) of the paper corresponding to the topic. At least a Powerpoint/PDF presentation and if possible some demonstration of an implementation or visualization has to be given. Teams are also expected to provide feedback on some of the presentations given by their fellow students during the lectures.
- Making a small contribution, i.e., doing something new compared to the paper on which the project is based. For example, a new tweak to an existing algorithm, a large number of datasets to test the algorithms to find a relation between the network characteristics and the performance, a new performance metric to evaluatie the algorithms, a new type of visualization of the algorithm or results, an improvement of a proof related to the algorithm, etc. In case of doubt about the contribution, contact the course staff well in advance; you can always ask for help.
-
Gathering and implementing the algorithms and/or techniques from the different papers, and running experiments on at least five large real-world network datasets.
Teams will also give feedback on the code produced by other teams in the code review session.
Some papers introduce multiple techniques. In that case, choose a logical subset to compare, and motivate your choice.
Datasets can for example be found at SNAP, KONECT, BigDND, Networks Repository, ICON, Netzschleuder, LASAGNE and ASU. Certain topics need particular datasets (e.g. with timestamps, signed links, etc.), which should of course be taken into account when selecting datasets. - Writing one 6 to 10 page paper. In the paper the different techniques are analyzed and compared in detail using extensive experiments. The paper, to be written in LaTeX, has to follow the format of an actual scientific paper. Students will also give feedback on the paper produced by another team in the peer review session.
Instructions for the course project paper are available.
Project topics
This list of projects is shown below. Please choose a topic with your team (consisting of two students). Register your choice in Brightspace. The group registration on Brightspace opens September 18 at 11:30. First come, first serve. Please choose a topic from the category that you have earlier joined on Brightspace: on-site (A) or online (C), or from the category from which all students can choose (B).
A. On-site group
- Anomaly detection 2
- Closeness centrality 2 (top-k)
- Community detection 4
- Data errors in networks 2
- Dense subgraph detection
- Graph evolution rules
- Link prediction in signed networks
- Link prediction 2
- Local cluster detection
- Motifs in temporal networks
- Network embeddings 1
- Privacy and anonymity in networks
- Sampling from networks 2
- Shortest paths 1
- Visualization algorithms 1
B. Online or on-site group
- Betweenness centrality 2
- Community detection 3
- Graph compression
- Graph obfuscation
- Influence spread and virality 1
- Motifs in networks
- Neighborhood approximation
- Personalized PageRank
- Triangle counting
- Visualization algorithms 3
C. Online group
- Anomaly detection 1
- Betweenness centrality 1
- Closeness centrality 1
- Community detection 1
- Community detection 2
- Data errors in networks 1
- Diameter computation
- Influence spread and virality 2
- Link prediction 1
- Motifs in multilayer networks
- Network embeddings 2
- Sampling from networks 1
- Shortest paths 2
- Visualization algorithms 2
Note: scientific papers (ACM, Elsevier, etc.) can often only be opened from within the university domain (or from home via SSH/VPN/etc.). IEEE Explore papers can often be opened by looking them up via computer.org. Alternative links and preprints of papers can often be found through Google Scholar by searching for "Title of the paper". Contact course staff if you have tried all of these options and are still not able to access the paper (do not pay!).
Reading material
Some students have expressed interest in additional reading material to help freshen up on skills and knowledge required for this course.
- To brush up on your Python programming skills, you could do Lecture 0, 1 and 4 of these Scientific Python Lectures.
- To freshen up your LaTeX skills, you could do for example this tutorial.
- MIT has a nice online course on discrete mathematics. Relevant chapters: 5, 6 and 7.
- The (Dutch) Leiden courses Algoritmiek (Algorithms) and Datastructuren (Data structures) might be also interesting to browse through.
- Basic knowledge of the concepts in machine learning and data mining are provided, for example, at the Wikipedia machine learning page.
Course description
See the e-Studyguide for a more general description.
Topics include: SNA from a CS perspective (graph representation, complexity issues, examples), Graph Structure (power law, small world phenomenon, clustering coefficient, hierarchies), Paths and Distances (neighborhoods, radius, diameter), Spidering and Sampling (BFS, forest fire, random walks), Graph Compression (graph grammars, bitwise tricks, encryption, hashing), Centrality (degree centrality, closeness centrality, betweenness centrality, rating and ranking), Centrality and Webgraphs (HITS, PageRank, structure of the web), Community Detection (spectral clustering, modularity), Visualization (force-based algorithms, Gephi, NodeXL), Graph Models (random graphs, preferential attachment), Link Prediction (structure, semantics, prediction algorithms, graph mining), Contagion (diffusion of information, spreading activation, gossipping) and Privacy and Anonymity ((de-)anonymizing graphs, ethical aspects, privacy issues) and various other topics that have been added over the years but are not yet in the list above.
The course was also given in 2014, 2015, 2016, 2017, 2018 and 2019.